Difference Between Mismatch Repair and Nucleotide Excision Repair
Table of Contents
Key Difference – Mismatch Repair vs Nucleotide Excision Repair
Tens and thousands of DNA damages occur in the cell per day. It induces changes to the cell processes such as replication, transcription as well as the viability of the cell. In some cases, mutations caused by these DNA damages may lead to deleterious diseases like cancers and aging-associated syndromes (ex: Progeria). Regardless of these damages, the cell initiates a highly organized cascade repair mechanism called DNA damage responses. Several DNA repair systems have been identified in the cellular system; these are known as Base excision repair (BER), Mismatch repair (MMR), Nucleotide excision repair (NER), Double strand break repair. Nucleotide excision repair is a highly versatile system that recognizes bulky helix distortion DNA lesions and removes them. On the other hand, mismatch repair replaces misincorporated bases during replication. The key difference between mismatch repair and nucleotide excision repair is that nucleotide excision repair (NER) is used to remove pyrimidine dimers formed by UV irradiation and bulky helix lesions caused by chemical adducts while mismatch repair system plays an important role in correcting misincorporated bases that have escaped from replication enzymes (DNA polymerase 1) during postreplication. In addition to mismatched bases, MMR system proteins can also repair the insertions/ deletions loops (IDL) which are results of the polymerase slippage during replication of repetitive DNA sequences.
CONTENTS
1. Overview and Key Difference
2. What is Mismatch Repair
3. What is Nucleotide Excision Repair
4. Side by Side Comparison – Mismatch Repair vs Nucleotide Excision Repair
5. Summary
What is Nucleotide Excision Repair?
The most distinguished feature of nucleotide excision repair is that it repairs the modified nucleotide damages caused by significant distortions in the DNA double helix. It is observed in almost all organisms that have been examined up to date. Uvr A, Uvr B, Uvr C (excinucleases) Uvr D (a helicase) are the best-known enzymes involved in the NER which trigger the repair of DNA in the model organism Ecoli. Uvr ABC multi-subunits enzyme complex produces the Uvr A, Uvr B, Uvr C polypeptides. The genes encoded for aforementioned polypeptides are uvr A, uvr B, uvr C. Uvr A and B enzymes collectively recognize the damage induced distortion that is caused to the DNA double helix such as pyrimidine dimmers due to UV irradiation. Uvr A is an ATPase enzyme and this is an autocatalytic reaction. Then Uvr A leaves the DNA whereas Uvr BC complex (active nuclease) cleave the DNA in both sides of the damage that catalyzed by ATP. Another protein called Uvr D encoded by uvrD gene is a helicase II enzyme unwinds the DNA that results from the releasing of single stranded damaged DNA segment. This leaves a gap in the DNA helix. After damaged segment has been excised, a 12-13 nucleotide gap remains in the DNA strand. This is filled up by the DNA polymerase enzyme I and the nick is sealed by the DNA ligase. ATP is required at three steps of this reaction. The NER mechanism can be identified in the mammalian-like humans as well. In humans, the skin condition called Xeroderma pigmentosum is due to the DNA dimers caused by UV irradiation. The genes XPA, XPB, XPC, XPD, XPE, XPF, and XPG produce proteins to replace DNA damage. The proteins of genes XPA, XPC, XPE, XPF, and XPG have the nuclease activity. On the other hand, the proteins of XPB and XPD genes show the helicase activity which analogs to Uvr D in E coli.
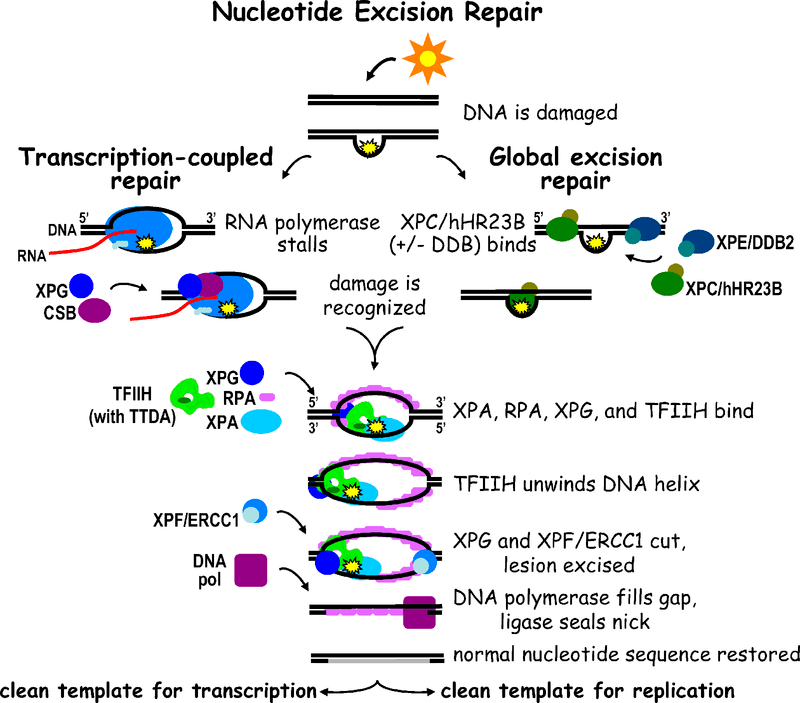
Figure 01: Nucleotide Excision repair
What is Mismatch Repair?
The mismatch repair system is initiated during DNA synthesis. Even with the functional € subunit, DNA polymerase III allows the incorporation of a wrong nucleotide for the synthesis every 108 base pairs. Mismatch repair proteins recognize this nucleotide, excise it and replace it with the correct nucleotide responsible for the final degree of accuracy. DNA methylation is pivotal for MMR proteins to recognize the parent strand from the newly synthesized strand. The methylation of adenine (A) nucleotide in a GATC motif of a newly synthesized strand is a little delayed. On the other hand, the parent strand adenine nucleotide in GATC motif has already methylated. MMR proteins recognize the newly synthesized strand by this difference from the parent strand and start mismatch repair in a newly synthesized strand before it gets methylated. The MMR proteins direct their repair activity to excise the wrong nucleotide before the newly replicated DNA strand gets methylated. The enzymes Mut H, Mut L and Mut S encoded by genes mut H, mut L, mut S catalyze these reactions in Ecoli. Mut S protein recognizes seven out of eight possible mismatch base pairs except for C:C, and binds at the site of mismatch in the duplex DNA. With bound ATPs, Mut L and Mut S join the complex later. The complex translocates few thousand base pairs away till it finds a hemimethylated GATC motif. The dormant nuclease activity of Mut H protein is activated once it finds a hemimethylated GATC motif. It cleaves the unmethylated DNA strand leaving a 5′ nick at G nucleotide of unmethylated GATC motif (newly synthesized DNA strand). Then the same strand on the other side of the mismatch is nicked by Mut H. In the rest of the steps, the collective actions of Uvr D a helicase protein, Mut U, SSB and exonuclease I excise the incorrect nucleotide in the single-stranded DNA. The gap which is formed in the excision is filled up by the DNA polymerase III and sealed by ligase. A similar system can be identified in mice and humans. The mutation of human hMLH1, hMSH1, and hMSH2 are involved in hereditary nonpolyposis colon cancer which deregulates the cell division of colon cells.
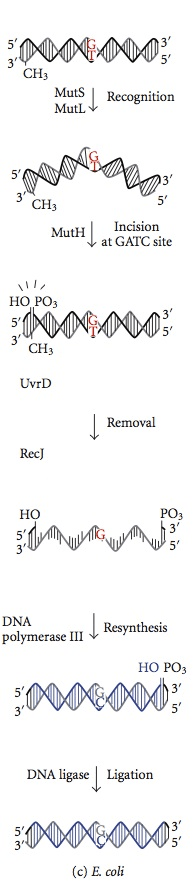
Figure 02: Mismatch Repair
What is the difference between Mismatch Repair and Nucleotide Excision Repair?
Mismatch Repair vs Nucleotide Excision Repair | |
| Mismatch repair system occurs during the post-replication. | This is involved in removing pyrimidine dimers due to U.V irradiation & other DNA lesions due to chemical adduct. |
| Enzymes | |
| It is catalyzed by Mut S, Mut L, Mut H, Uvr D, SSB and exonuclease I. | It is catalyzed by Uvr A, Uvr B, Uvr C, UvrD enzymes. |
| Methylation | |
| It is pivotal to initiate the reaction. | DNA methylation is not required for initiating the reaction. |
| Action of Enzymes | |
| Mut H is an endonuclease. | Uvr B and Uvr C are exonucleases. |
| Occasion | |
| This happens specifically during replication. | This happens when exposed to U.V or chemical mutagens, not during replication |
| Conservation | |
| It is highly conserved | It is not highly conserved. |
| Gap Filling | |
| It is done by DNA polymerase III. | It is done by DNA polymerase I. |
Summary – Mismatch Repair vs Nucleotide Excision Repair
Mismatch repair (MMR) and Nucleotide excision repair (NER) are two mechanisms that take place in the cell in order to rectify DNA damages and distortions that are caused by various agents. These are collectively named as DNA repair mechanisms. Nucleotide excision repair repairs the modified nucleotide damages, typically those significant damages of the DNA double helix which happen due to exposure to U.V irradiation and chemical adducts. Mismatch repair proteins recognize the wrong nucleotide, excise it and replace it with correct nucleotide. This process is responsible for the final degree of accuracy during replication.
Reference:
1.Cooper, Geoffrey M. “DNA Repair.” The Cell: A Molecular Approach. 2nd edition.U.S. National Library of Medicine, 01 Jan. 1970. Web. 09 Mar. 2017.
2.”Mechanisms and functions of DNA mismatch repair.” Cell research. U.S. National Library of Medicine, n.d. Web. 09 Mar. 2017.
Image Courtesy:
1. “Nucleotide Excision Repair-journal.pbio.0040203.g001” By Jill O. Fuss, Priscilla K. Cooper – (CC BY 2.5) via Commons Wikimedia
2. “DNA mismatch repair Ecoli” By Kenji Fukui – (CC BY 4.0) via Commons Wikimedia
ncG1vNJzZmivp6x7pbXFn5yrnZ6YsqOx07CcnqZemLyue8OinZ%2Bdopq7pLGMm5ytr5Wau265yKykmqyTnXqzsc%2BaoKtlkaOxbsLSZqWum5yavLW1w55knrCTnsCqu81mqZ6okZ6%2FcA%3D%3D