Difference Between Maxam Gilbert and Sanger Sequencing
Table of Contents
Key Difference – Maxam Gilbert vs Sanger Sequencing
Nucleotides are the basic structural units and building blocks of DNA. DNA molecule is composed of a polynucleotide chain. There are four different nucleotides found in DNA. These nucleotides are composed of four different nitrogenous bases named A (adenine), G (guanine), C (cytosine), T (thymine). The order of the nucleotides in the DNA molecule has a great importance as it encodes an important genetic information for organisms growth and development. DNA sequencing refers to the process which determines the precise nucleotide sequence of the DNA. There are different DNA sequencing methods. Maxam Gilbert sequencing and Sanger DNA sequencing are two methods of DNA sequencing which belong to first generation sequencing. Maxam Gilbert sequencing procedure determines the base sequence by chemically cleaving the 5’ end labelled DNA fragments preferentially at each of four nucleotides and gel electrophoresis. Sanger sequencing procedure determines the nucleotide sequence by synthesizing single-stranded DNA using DNA polymerase and dideoxynucleotides and gel electrophoresis. This is the key difference between Maxam Gilbert and Sanger Sequencing.
CONTENTS
1. Overview and Key Difference
2. What is Maxam Gilbert
3. What is Sanger Sequencing
4. Side by Side Comparison – Maxam Gilbert vs Sanger Sequencing
5. Summary
What is Maxam Gilbert Sequencing?
Maxam Gilbert sequencing, also known as chemical sequencing method, is a technique which was developed to determine the order of the nucleotides in DNA. This method was introduced by Walter Gilbert and Alan Maxam in 1976 and became popular since it can be performed directly with purified DNA. Maxam Gilbert method belongs to the first generation of DNA sequencing, and it was the first sequencing method used widely by scientists.
The basic principle of this method lies with the restriction of the end-labeled DNA fragments at specific bases by base-specific chemicals and conditions and separation of the labeled fragments by electrophoresis as shown in figure 01. Fragments are separated according to their sizes on the gel. Since the fragments are labeled, the sequence of the DNA molecule can be easily deduced.
Maxam gilbert method uses base specific chemicals to break DNA at specific bases. Two common chemicals named dimethyl sulphate and hydrazine chemicals are used to selectively attack purines and pyrimidines, respectively.
Maxam Gilbert sequencing method is performed via several steps as follows.
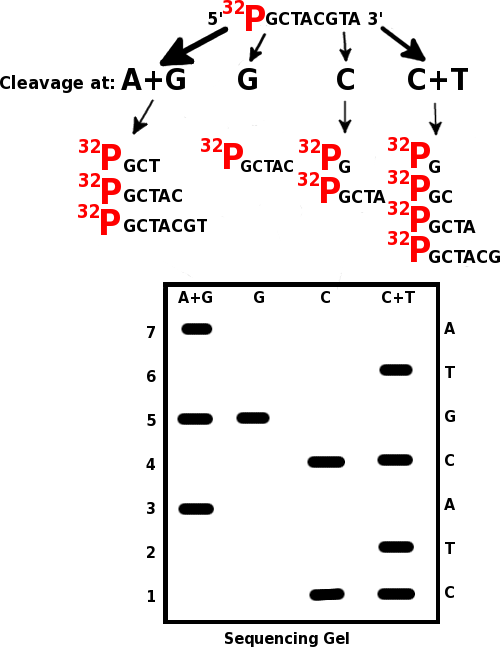
Figure 01: Maxam Gilbert Sequencing
What is Sanger Sequencing?
Sanger Sequencing is a sequencing method developed by Frederick Sanger and his colleagues in 1977 to determine the base sequence of a given DNA fragment. It is also known as chain termination sequencing or Dideoxy sequencing method. This method works on the principle of selective incorporation of chain terminating dideoxynucleotides (ddNTPs) such as ddGTP, ddCTP, ddATP and ddTTP by DNA polymerase during the synthesis of single stranded DNA to terminate strand formation. Dideoxynucleotides lack 3’ OH groups for the formation of phosphodiester bonds with adjacent nucleotide. Hence, the strand formation stops once a ddNTP is incorporated into newly forming strand during sanger sequencing.
In this method, four separate DNA synthesis reactions (PCR) are performed in four separate tubes with one type of ddNTP. Other requirements are also supplied for the tubes for PCR including primers, dNTPs, Taq polymerase, specific conditions, etc. Four separate reactions are performed in four tubes with four mixtures. After PCR reactions, resulting DNA fragments are heat denatured and separated by gel electrophoresis. Then the fragments are visualized by using either a labeled (radioactive or fluorescent) primer or dNTPs as shown in figure 02.

Figure 02: Sanger Sequencing
What is the difference between Maxam Gilbert and Sanger Sequencing?
Maxam Gilbert vs Sanger Sequencing | |
| Maxam Gilbert sequencing is the first technique developed for DNA sequencing. | Sanger sequencing method was introduced after the Maxam Gilbert sequencing method. |
| Usage | |
| This method is rarely used. | Sanger sequencing is routinely used for sequencing. |
| Use of Hazardous Chemicals | |
| It uses hazardous chemicals. | Use of hazardous chemicals is limited compared to Maxam Gilbert method. |
| Labeling for Detection | |
| This method uses radioactive P32 for labeling the ends of the DNA fragments. | Sanger sequencing uses radioactively or fluorescently labeled ddNTPs. |
Summary – Maxam Gilbert vs Sanger Sequencing
Maxam Gilbert and Sanger sequencing are two types of DNA sequencing techniques coming under first generation DNA sequencing. Maxam Gilbert sequencing is the first method introduced for DNA sequencing in 1976, and it is performed by breaking the end labeled DNA fragments by base-specific chemicals. Hence, it is known as chemical sequencing. Sanger sequencing method was introduced in 1977, and is based on the ddNTP driven chain termination reactions. Sanger sequencing method popular than Maxam Gilbert method due to several disadvantages of Maxam Gilbert method such as excessive time consumption, use of hazardous chemicals, etc. This is the difference between Maxam Gilbert and Sanger sequencing.
References:
1.Maxam, A. M, and Walter Gilbert. “A new method for sequencing DNA.”Proceedings of the National Academy of Sciences. National Acad Sciences, 9 Dec. 1976. Web. 30 Mar. 2017
2.Heather, James M., and Benjamin Chain. “The sequence of sequencers: The history of sequencing DNA.” Genomics. Academic Press, Jan. 2016. Web. 30 Mar. 2017
3.Pareek, Chandra Shekhar, Rafal Smoczynski, and Andrzej Tretyn. “Sequencing technologies and genome sequencing.” Journal of Applied Genetics. Springer-Verlag, Nov. 2011. Web. 30 Mar. 2017
Image Courtesy:
1. “Maxam gilbert sequencing” By Several Times at English Wikipedia (CC BY 3.0) via Commons Wikimedia
2. “Sanger-sequencing” By Estevezj – Own work (CC BY-SA 3.0) via Commons Wikimedia
ncG1vNJzZmivp6x7pbXFn5yrnZ6YsqOx07CcnqZemLyue8OinZ%2Bdopq7pLGMm5ytr5Wau265wLGYpmWXnrmjsdGtZJqmlGLDtHnSmqWgnaJiwKa91J6lnKGenHw%3D